Session details
Objectives
- To learn the difference between “messy” and “tidy” data and see the advantages of using data that are consistently structured.
- To perform simple data transformations and create summaries.
- To get to know the tools that help tidying up and reshaping the data.
At the end of this session you will be able:
- To write code using the
%>%operator. - To perform simple data transformations using
mutate(). - To select variables and observations to work with using
filter()andselect(). - To order data by variable using
arrange(). - To provide a simple data summary using
group_by()andsummarise(). - To change the structure of the data using
gather()(and optionallyspread()).
Expected learning
For the final exercise of this session, you will try to get the data from the original data (using NHANES) to the table below. By using the code in this session, you will have acheived our learning expectations.
The final exercise will be to get this data:
NHANES
#> # A tibble: 10,000 x 76
#> ID SurveyYr Gender Age AgeDecade AgeMonths Race1 Race3 Education
#> <int> <fct> <fct> <int> <fct> <int> <fct> <fct> <fct>
#> 1 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 2 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 3 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 4 51625 2009_10 male 4 " 0-9" 49 Other <NA> <NA>
#> 5 51630 2009_10 female 49 " 40-49" 596 White <NA> Some Col…
#> 6 51638 2009_10 male 9 " 0-9" 115 White <NA> <NA>
#> 7 51646 2009_10 male 8 " 0-9" 101 White <NA> <NA>
#> 8 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 9 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 10 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> # … with 9,990 more rows, and 67 more variables: MaritalStatus <fct>,
#> # HHIncome <fct>, HHIncomeMid <int>, Poverty <dbl>, HomeRooms <int>,
#> # HomeOwn <fct>, Work <fct>, Weight <dbl>, Length <dbl>, HeadCirc <dbl>,
#> # Height <dbl>, BMI <dbl>, BMICatUnder20yrs <fct>, BMI_WHO <fct>,
#> # Pulse <int>, BPSysAve <int>, BPDiaAve <int>, BPSys1 <int>,
#> # BPDia1 <int>, BPSys2 <int>, BPDia2 <int>, BPSys3 <int>, BPDia3 <int>,
#> # Testosterone <dbl>, DirectChol <dbl>, TotChol <dbl>, UrineVol1 <int>,
#> # UrineFlow1 <dbl>, UrineVol2 <int>, UrineFlow2 <dbl>, Diabetes <fct>,
#> # DiabetesAge <int>, HealthGen <fct>, DaysPhysHlthBad <int>,
#> # DaysMentHlthBad <int>, LittleInterest <fct>, Depressed <fct>,
#> # nPregnancies <int>, nBabies <int>, Age1stBaby <int>,
#> # SleepHrsNight <int>, SleepTrouble <fct>, PhysActive <fct>,
#> # PhysActiveDays <int>, TVHrsDay <fct>, CompHrsDay <fct>,
#> # TVHrsDayChild <int>, CompHrsDayChild <int>, Alcohol12PlusYr <fct>,
#> # AlcoholDay <int>, AlcoholYear <int>, SmokeNow <fct>, Smoke100 <fct>,
#> # Smoke100n <fct>, SmokeAge <int>, Marijuana <fct>, AgeFirstMarij <int>,
#> # RegularMarij <fct>, AgeRegMarij <int>, HardDrugs <fct>, SexEver <fct>,
#> # SexAge <int>, SexNumPartnLife <int>, SexNumPartYear <int>,
#> # SameSex <fct>, SexOrientation <fct>, PregnantNow <fct>
To look like this:
| Gender | Measure | 2009_10 | 2011_12 |
|---|---|---|---|
| female | Age | 44.02 | 44.15 |
| female | AgeDiabetesDiagnosis | 48.13 | 46.51 |
| female | BMI | 28.96 | 28.60 |
| female | BPDiaAve | 67.69 | 70.04 |
| female | BPSysAve | 116.04 | 117.90 |
| female | DrinksOfAlcoholInDay | 2.34 | 2.20 |
| female | MoreThan5DaysActive | 0.31 | 0.39 |
| female | NumberOfBabies | 2.44 | 2.31 |
| female | Poverty | 2.93 | 2.88 |
| female | TotalCholesterol | 5.14 | 5.11 |
| male | Age | 43.11 | 43.88 |
| male | AgeDiabetesDiagnosis | 47.58 | 45.14 |
| male | BMI | 28.76 | 28.70 |
| male | BPDiaAve | 70.94 | 73.14 |
| male | BPSysAve | 121.61 | 122.43 |
| male | DrinksOfAlcoholInDay | 3.52 | 3.71 |
| male | MoreThan5DaysActive | 0.29 | 0.36 |
| male | NumberOfBabies | NaN | NaN |
| male | Poverty | 3.01 | 2.97 |
| male | TotalCholesterol | 5.07 | 4.90 |
Managing and working with data in R
Recall that:
- You should never edit your raw data (should be in
data-raw/folder) - Only work with your raw data using R code
- Save the edited data as another dataset (should be in
data/folder) - Document and comment as best you can to help you remember
- But, let the code speak for itself (e.g. keep it simple, have intermediate and descriptive steps)
- But, let the code speak for itself (e.g. keep it simple, have intermediate and descriptive steps)
In data “wrangling”/“munging”/managing, most tasks can be broken down into only a few simple “verbs” (actions), as listed in the table.
| Tasks | Examples | Function |
|---|---|---|
| Choosing columns | Remove columns related to data entry, such as name of person entering data. | select() |
| Renaming columns | Changing a column name from 'Q1' to 'ParticipantName'. | rename() |
| Transforming or modifying columns | Multiplying a column's values; taking the log. | mutate() |
| Subsetting/filtering out rows/observations | Keeping rows with glucose values above 4. | filter() |
| Sorting/re-arranging rows | Show rows with the smallest value at the top. | arrange() |
| Converting from wide to long form | One row per participant to multiple participants per row (repeated measures). | gather() |
| Converting from long to wide form | Multiple rows per participant (repeated measures) to one participant per row. | spread() |
| Calculating summaries on the data | Calculating the maximum, median, and minimum age. | summarise() |
| Running analyses by group | Calculate means of age by males and females. | group_by() with summarise() |
Wrangling here is used in the sense of maneuvering, managing, controlling, and turning your data around to better understand it and to prepare it for later analyses. The packages dplyr and tidyr provide easy tools for most common data manipulation tasks. For dplyr, it is built to work directly with data frames and has an additional feature to interact directly with data stored in an external database, such as in SQL. Working with databases is powerful as you can work with massive datasets (100s of GB), more than your computer could normally handle. This won’t be covered, but see this resources from Data Carpentry) to learn more.
A tip when doing complicated wrangling and you get the data to a form that you
like and will use often: Save it as an “output” dataset in the data/ folder so
you can easily use it again later rather than run the wrangling code everytime.
Loading the packages and dataset
We’re going to use the US NHANES dataset. There is an NHANES package that
contains a teaching version of the original dataset, so we’ll use that for this
lesson. First, make sure the R Project you created previously is open. Then open
the R/wrangling-session.R script to start typing out the next code. We’ll use
this file to write the code for this session (but not for the exercises).
# Load up the packages
# tidyverse contains the dplyr and tidyr packages
library(tidyverse)
library(NHANES)
# Briefly glimpse contents of dataset
glimpse(NHANES)
#> Observations: 10,000
#> Variables: 76
#> $ ID <int> 51624, 51624, 51624, 51625, 51630, 51638, 51646…
#> $ SurveyYr <fct> 2009_10, 2009_10, 2009_10, 2009_10, 2009_10, 20…
#> $ Gender <fct> male, male, male, male, female, male, male, fem…
#> $ Age <int> 34, 34, 34, 4, 49, 9, 8, 45, 45, 45, 66, 58, 54…
#> $ AgeDecade <fct> 30-39, 30-39, 30-39, 0-9, 40-49, 0-9, 0-…
#> $ AgeMonths <int> 409, 409, 409, 49, 596, 115, 101, 541, 541, 541…
#> $ Race1 <fct> White, White, White, Other, White, White, White…
#> $ Race3 <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
#> $ Education <fct> High School, High School, High School, NA, Some…
#> $ MaritalStatus <fct> Married, Married, Married, NA, LivePartner, NA,…
#> $ HHIncome <fct> 25000-34999, 25000-34999, 25000-34999, 20000-24…
#> $ HHIncomeMid <int> 30000, 30000, 30000, 22500, 40000, 87500, 60000…
#> $ Poverty <dbl> 1.36, 1.36, 1.36, 1.07, 1.91, 1.84, 2.33, 5.00,…
#> $ HomeRooms <int> 6, 6, 6, 9, 5, 6, 7, 6, 6, 6, 5, 10, 6, 10, 10,…
#> $ HomeOwn <fct> Own, Own, Own, Own, Rent, Rent, Own, Own, Own, …
#> $ Work <fct> NotWorking, NotWorking, NotWorking, NA, NotWork…
#> $ Weight <dbl> 87.4, 87.4, 87.4, 17.0, 86.7, 29.8, 35.2, 75.7,…
#> $ Length <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
#> $ HeadCirc <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
#> $ Height <dbl> 164.7, 164.7, 164.7, 105.4, 168.4, 133.1, 130.6…
#> $ BMI <dbl> 32.22, 32.22, 32.22, 15.30, 30.57, 16.82, 20.64…
#> $ BMICatUnder20yrs <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
#> $ BMI_WHO <fct> 30.0_plus, 30.0_plus, 30.0_plus, 12.0_18.5, 30.…
#> $ Pulse <int> 70, 70, 70, NA, 86, 82, 72, 62, 62, 62, 60, 62,…
#> $ BPSysAve <int> 113, 113, 113, NA, 112, 86, 107, 118, 118, 118,…
#> $ BPDiaAve <int> 85, 85, 85, NA, 75, 47, 37, 64, 64, 64, 63, 74,…
#> $ BPSys1 <int> 114, 114, 114, NA, 118, 84, 114, 106, 106, 106,…
#> $ BPDia1 <int> 88, 88, 88, NA, 82, 50, 46, 62, 62, 62, 64, 76,…
#> $ BPSys2 <int> 114, 114, 114, NA, 108, 84, 108, 118, 118, 118,…
#> $ BPDia2 <int> 88, 88, 88, NA, 74, 50, 36, 68, 68, 68, 62, 72,…
#> $ BPSys3 <int> 112, 112, 112, NA, 116, 88, 106, 118, 118, 118,…
#> $ BPDia3 <int> 82, 82, 82, NA, 76, 44, 38, 60, 60, 60, 64, 76,…
#> $ Testosterone <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
#> $ DirectChol <dbl> 1.29, 1.29, 1.29, NA, 1.16, 1.34, 1.55, 2.12, 2…
#> $ TotChol <dbl> 3.49, 3.49, 3.49, NA, 6.70, 4.86, 4.09, 5.82, 5…
#> $ UrineVol1 <int> 352, 352, 352, NA, 77, 123, 238, 106, 106, 106,…
#> $ UrineFlow1 <dbl> NA, NA, NA, NA, 0.094, 1.538, 1.322, 1.116, 1.1…
#> $ UrineVol2 <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
#> $ UrineFlow2 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
#> $ Diabetes <fct> No, No, No, No, No, No, No, No, No, No, No, No,…
#> $ DiabetesAge <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
#> $ HealthGen <fct> Good, Good, Good, NA, Good, NA, NA, Vgood, Vgoo…
#> $ DaysPhysHlthBad <int> 0, 0, 0, NA, 0, NA, NA, 0, 0, 0, 10, 0, 4, NA, …
#> $ DaysMentHlthBad <int> 15, 15, 15, NA, 10, NA, NA, 3, 3, 3, 0, 0, 0, N…
#> $ LittleInterest <fct> Most, Most, Most, NA, Several, NA, NA, None, No…
#> $ Depressed <fct> Several, Several, Several, NA, Several, NA, NA,…
#> $ nPregnancies <int> NA, NA, NA, NA, 2, NA, NA, 1, 1, 1, NA, NA, NA,…
#> $ nBabies <int> NA, NA, NA, NA, 2, NA, NA, NA, NA, NA, NA, NA, …
#> $ Age1stBaby <int> NA, NA, NA, NA, 27, NA, NA, NA, NA, NA, NA, NA,…
#> $ SleepHrsNight <int> 4, 4, 4, NA, 8, NA, NA, 8, 8, 8, 7, 5, 4, NA, 5…
#> $ SleepTrouble <fct> Yes, Yes, Yes, NA, Yes, NA, NA, No, No, No, No,…
#> $ PhysActive <fct> No, No, No, NA, No, NA, NA, Yes, Yes, Yes, Yes,…
#> $ PhysActiveDays <int> NA, NA, NA, NA, NA, NA, NA, 5, 5, 5, 7, 5, 1, N…
#> $ TVHrsDay <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
#> $ CompHrsDay <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
#> $ TVHrsDayChild <int> NA, NA, NA, 4, NA, 5, 1, NA, NA, NA, NA, NA, NA…
#> $ CompHrsDayChild <int> NA, NA, NA, 1, NA, 0, 6, NA, NA, NA, NA, NA, NA…
#> $ Alcohol12PlusYr <fct> Yes, Yes, Yes, NA, Yes, NA, NA, Yes, Yes, Yes, …
#> $ AlcoholDay <int> NA, NA, NA, NA, 2, NA, NA, 3, 3, 3, 1, 2, 6, NA…
#> $ AlcoholYear <int> 0, 0, 0, NA, 20, NA, NA, 52, 52, 52, 100, 104, …
#> $ SmokeNow <fct> No, No, No, NA, Yes, NA, NA, NA, NA, NA, No, NA…
#> $ Smoke100 <fct> Yes, Yes, Yes, NA, Yes, NA, NA, No, No, No, Yes…
#> $ Smoke100n <fct> Smoker, Smoker, Smoker, NA, Smoker, NA, NA, Non…
#> $ SmokeAge <int> 18, 18, 18, NA, 38, NA, NA, NA, NA, NA, 13, NA,…
#> $ Marijuana <fct> Yes, Yes, Yes, NA, Yes, NA, NA, Yes, Yes, Yes, …
#> $ AgeFirstMarij <int> 17, 17, 17, NA, 18, NA, NA, 13, 13, 13, NA, 19,…
#> $ RegularMarij <fct> No, No, No, NA, No, NA, NA, No, No, No, NA, Yes…
#> $ AgeRegMarij <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 20,…
#> $ HardDrugs <fct> Yes, Yes, Yes, NA, Yes, NA, NA, No, No, No, No,…
#> $ SexEver <fct> Yes, Yes, Yes, NA, Yes, NA, NA, Yes, Yes, Yes, …
#> $ SexAge <int> 16, 16, 16, NA, 12, NA, NA, 13, 13, 13, 17, 22,…
#> $ SexNumPartnLife <int> 8, 8, 8, NA, 10, NA, NA, 20, 20, 20, 15, 7, 100…
#> $ SexNumPartYear <int> 1, 1, 1, NA, 1, NA, NA, 0, 0, 0, NA, 1, 1, NA, …
#> $ SameSex <fct> No, No, No, NA, Yes, NA, NA, Yes, Yes, Yes, No,…
#> $ SexOrientation <fct> Heterosexual, Heterosexual, Heterosexual, NA, H…
#> $ PregnantNow <fct> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
Exercise: Become familiar with the dataset
Time: 10 min
Take the time to get familiar with the NHANES dataset. Create a new R script
by typing in the console usethis::use_r("exercises-wrangling"). Then copy the
code below into the file and replace the ___ with the NHANES dataset. Run
each line of code by typing Ctrl-Enter.
# Load the packages
library(tidyverse)
library(NHANES)
# Check column names
colnames(___)
# Look at contents
str(___)
glimpse(___)
# See summary
summary(___)
# Look over the dataset documentation
?___
Click for a possible solution
# load the packages
library(tidyverse)
library(NHANES)
# Check column names
colnames(NHANES)
# Look at contents
str(NHANES)
glimpse(NHANES)
# See summary
summary(NHANES)
# Look over the dataset documentation
?NHANES
“Messy” vs. “tidy” data
The concept of “tidy” data was popularized in an article by Hadley Wickham and described more in the Tidy Data chapter of the R for Data Science online book. A tidy dataset is when:
- Each variable has its own column.
- Each observation has its own row
- Each value has its own cell
Looking at these example datasets, think about why each is “tidy” or “messy”. What do you notice between the tidy versions and the messier versions?
# Datasets come from tidyr
# Tidy:
table1
#> # A tibble: 6 x 4
#> country year cases population
#> <chr> <int> <int> <int>
#> 1 Afghanistan 1999 745 19987071
#> 2 Afghanistan 2000 2666 20595360
#> 3 Brazil 1999 37737 172006362
#> 4 Brazil 2000 80488 174504898
#> 5 China 1999 212258 1272915272
#> 6 China 2000 213766 1280428583
# Tidy:
table2
#> # A tibble: 12 x 4
#> country year type count
#> <chr> <int> <chr> <int>
#> 1 Afghanistan 1999 cases 745
#> 2 Afghanistan 1999 population 19987071
#> 3 Afghanistan 2000 cases 2666
#> 4 Afghanistan 2000 population 20595360
#> 5 Brazil 1999 cases 37737
#> 6 Brazil 1999 population 172006362
#> 7 Brazil 2000 cases 80488
#> 8 Brazil 2000 population 174504898
#> 9 China 1999 cases 212258
#> 10 China 1999 population 1272915272
#> 11 China 2000 cases 213766
#> 12 China 2000 population 1280428583
# Messier:
table3
#> # A tibble: 6 x 3
#> country year rate
#> * <chr> <int> <chr>
#> 1 Afghanistan 1999 745/19987071
#> 2 Afghanistan 2000 2666/20595360
#> 3 Brazil 1999 37737/172006362
#> 4 Brazil 2000 80488/174504898
#> 5 China 1999 212258/1272915272
#> 6 China 2000 213766/1280428583
# Messy:
table4a
#> # A tibble: 3 x 3
#> country `1999` `2000`
#> * <chr> <int> <int>
#> 1 Afghanistan 745 2666
#> 2 Brazil 37737 80488
#> 3 China 212258 213766
# Messy:
table4b
#> # A tibble: 3 x 3
#> country `1999` `2000`
#> * <chr> <int> <int>
#> 1 Afghanistan 19987071 20595360
#> 2 Brazil 172006362 174504898
#> 3 China 1272915272 1280428583
In the tidy versions, table1 and table2 have columns that describe their values
(e.g. population is population size), each row is unique (e.g. first row is for
values from Afghanistan from 1999), and each cell is an explicit value
representative of its column and row. On the other hand, table3 is not tidy
because the rate column values are a composite of two other column values
(cases and population), when it should be a single number (a percent). Both
table4a and table4b have columns with ambiguous values inside… what does
values in the 1999 column contain? You can’t tell from the data.
Tidy data has a few notable benefits:
- Time spent preparing your data to be tidy from the beginning can save days of frustration in the long run.
- “Tidy data” is a conceptual framework that allows you to easy build off and wrangle data in simpler and easy to interpret ways, especially when using the tidyverse packages.
From the concept of tidy data also comes the concept of tidy code. By using “verbs” (R functions), chaining them together in “sentences” (in a sequential pipeline), you can construct meaningful and readable code that, in more plain English, describes what you are doing to the data.
%>%: The pipe operator
A key component of the tidy data and tidy code concept is the use of the %>%
operator. This operator allows you to “pipe” the output from one function to the
input of another function, thus allowing you to easily chain functions together
into “sentences”. So, instead of nesting functions (reading from the inside to
the outside), the idea of piping is to read the functions from left to right.
This can help clarify and break down complex data processing workflows.
# These two ways are the same
colnames(NHANES)
#> [1] "ID" "SurveyYr" "Gender"
#> [4] "Age" "AgeDecade" "AgeMonths"
#> [7] "Race1" "Race3" "Education"
#> [10] "MaritalStatus" "HHIncome" "HHIncomeMid"
#> [13] "Poverty" "HomeRooms" "HomeOwn"
#> [16] "Work" "Weight" "Length"
#> [19] "HeadCirc" "Height" "BMI"
#> [22] "BMICatUnder20yrs" "BMI_WHO" "Pulse"
#> [25] "BPSysAve" "BPDiaAve" "BPSys1"
#> [28] "BPDia1" "BPSys2" "BPDia2"
#> [31] "BPSys3" "BPDia3" "Testosterone"
#> [34] "DirectChol" "TotChol" "UrineVol1"
#> [37] "UrineFlow1" "UrineVol2" "UrineFlow2"
#> [40] "Diabetes" "DiabetesAge" "HealthGen"
#> [43] "DaysPhysHlthBad" "DaysMentHlthBad" "LittleInterest"
#> [46] "Depressed" "nPregnancies" "nBabies"
#> [49] "Age1stBaby" "SleepHrsNight" "SleepTrouble"
#> [52] "PhysActive" "PhysActiveDays" "TVHrsDay"
#> [55] "CompHrsDay" "TVHrsDayChild" "CompHrsDayChild"
#> [58] "Alcohol12PlusYr" "AlcoholDay" "AlcoholYear"
#> [61] "SmokeNow" "Smoke100" "Smoke100n"
#> [64] "SmokeAge" "Marijuana" "AgeFirstMarij"
#> [67] "RegularMarij" "AgeRegMarij" "HardDrugs"
#> [70] "SexEver" "SexAge" "SexNumPartnLife"
#> [73] "SexNumPartYear" "SameSex" "SexOrientation"
#> [76] "PregnantNow"
NHANES %>%
colnames()
#> [1] "ID" "SurveyYr" "Gender"
#> [4] "Age" "AgeDecade" "AgeMonths"
#> [7] "Race1" "Race3" "Education"
#> [10] "MaritalStatus" "HHIncome" "HHIncomeMid"
#> [13] "Poverty" "HomeRooms" "HomeOwn"
#> [16] "Work" "Weight" "Length"
#> [19] "HeadCirc" "Height" "BMI"
#> [22] "BMICatUnder20yrs" "BMI_WHO" "Pulse"
#> [25] "BPSysAve" "BPDiaAve" "BPSys1"
#> [28] "BPDia1" "BPSys2" "BPDia2"
#> [31] "BPSys3" "BPDia3" "Testosterone"
#> [34] "DirectChol" "TotChol" "UrineVol1"
#> [37] "UrineFlow1" "UrineVol2" "UrineFlow2"
#> [40] "Diabetes" "DiabetesAge" "HealthGen"
#> [43] "DaysPhysHlthBad" "DaysMentHlthBad" "LittleInterest"
#> [46] "Depressed" "nPregnancies" "nBabies"
#> [49] "Age1stBaby" "SleepHrsNight" "SleepTrouble"
#> [52] "PhysActive" "PhysActiveDays" "TVHrsDay"
#> [55] "CompHrsDay" "TVHrsDayChild" "CompHrsDayChild"
#> [58] "Alcohol12PlusYr" "AlcoholDay" "AlcoholYear"
#> [61] "SmokeNow" "Smoke100" "Smoke100n"
#> [64] "SmokeAge" "Marijuana" "AgeFirstMarij"
#> [67] "RegularMarij" "AgeRegMarij" "HardDrugs"
#> [70] "SexEver" "SexAge" "SexNumPartnLife"
#> [73] "SexNumPartYear" "SameSex" "SexOrientation"
#> [76] "PregnantNow"
# Standard R way of "chaining" functions together
glimpse(head(NHANES))
#> Observations: 6
#> Variables: 76
#> $ ID <int> 51624, 51624, 51624, 51625, 51630, 51638
#> $ SurveyYr <fct> 2009_10, 2009_10, 2009_10, 2009_10, 2009_10, 20…
#> $ Gender <fct> male, male, male, male, female, male
#> $ Age <int> 34, 34, 34, 4, 49, 9
#> $ AgeDecade <fct> 30-39, 30-39, 30-39, 0-9, 40-49, 0-9
#> $ AgeMonths <int> 409, 409, 409, 49, 596, 115
#> $ Race1 <fct> White, White, White, Other, White, White
#> $ Race3 <fct> NA, NA, NA, NA, NA, NA
#> $ Education <fct> High School, High School, High School, NA, Some…
#> $ MaritalStatus <fct> Married, Married, Married, NA, LivePartner, NA
#> $ HHIncome <fct> 25000-34999, 25000-34999, 25000-34999, 20000-24…
#> $ HHIncomeMid <int> 30000, 30000, 30000, 22500, 40000, 87500
#> $ Poverty <dbl> 1.36, 1.36, 1.36, 1.07, 1.91, 1.84
#> $ HomeRooms <int> 6, 6, 6, 9, 5, 6
#> $ HomeOwn <fct> Own, Own, Own, Own, Rent, Rent
#> $ Work <fct> NotWorking, NotWorking, NotWorking, NA, NotWork…
#> $ Weight <dbl> 87.4, 87.4, 87.4, 17.0, 86.7, 29.8
#> $ Length <dbl> NA, NA, NA, NA, NA, NA
#> $ HeadCirc <dbl> NA, NA, NA, NA, NA, NA
#> $ Height <dbl> 164.7, 164.7, 164.7, 105.4, 168.4, 133.1
#> $ BMI <dbl> 32.22, 32.22, 32.22, 15.30, 30.57, 16.82
#> $ BMICatUnder20yrs <fct> NA, NA, NA, NA, NA, NA
#> $ BMI_WHO <fct> 30.0_plus, 30.0_plus, 30.0_plus, 12.0_18.5, 30.…
#> $ Pulse <int> 70, 70, 70, NA, 86, 82
#> $ BPSysAve <int> 113, 113, 113, NA, 112, 86
#> $ BPDiaAve <int> 85, 85, 85, NA, 75, 47
#> $ BPSys1 <int> 114, 114, 114, NA, 118, 84
#> $ BPDia1 <int> 88, 88, 88, NA, 82, 50
#> $ BPSys2 <int> 114, 114, 114, NA, 108, 84
#> $ BPDia2 <int> 88, 88, 88, NA, 74, 50
#> $ BPSys3 <int> 112, 112, 112, NA, 116, 88
#> $ BPDia3 <int> 82, 82, 82, NA, 76, 44
#> $ Testosterone <dbl> NA, NA, NA, NA, NA, NA
#> $ DirectChol <dbl> 1.29, 1.29, 1.29, NA, 1.16, 1.34
#> $ TotChol <dbl> 3.49, 3.49, 3.49, NA, 6.70, 4.86
#> $ UrineVol1 <int> 352, 352, 352, NA, 77, 123
#> $ UrineFlow1 <dbl> NA, NA, NA, NA, 0.094, 1.538
#> $ UrineVol2 <int> NA, NA, NA, NA, NA, NA
#> $ UrineFlow2 <dbl> NA, NA, NA, NA, NA, NA
#> $ Diabetes <fct> No, No, No, No, No, No
#> $ DiabetesAge <int> NA, NA, NA, NA, NA, NA
#> $ HealthGen <fct> Good, Good, Good, NA, Good, NA
#> $ DaysPhysHlthBad <int> 0, 0, 0, NA, 0, NA
#> $ DaysMentHlthBad <int> 15, 15, 15, NA, 10, NA
#> $ LittleInterest <fct> Most, Most, Most, NA, Several, NA
#> $ Depressed <fct> Several, Several, Several, NA, Several, NA
#> $ nPregnancies <int> NA, NA, NA, NA, 2, NA
#> $ nBabies <int> NA, NA, NA, NA, 2, NA
#> $ Age1stBaby <int> NA, NA, NA, NA, 27, NA
#> $ SleepHrsNight <int> 4, 4, 4, NA, 8, NA
#> $ SleepTrouble <fct> Yes, Yes, Yes, NA, Yes, NA
#> $ PhysActive <fct> No, No, No, NA, No, NA
#> $ PhysActiveDays <int> NA, NA, NA, NA, NA, NA
#> $ TVHrsDay <fct> NA, NA, NA, NA, NA, NA
#> $ CompHrsDay <fct> NA, NA, NA, NA, NA, NA
#> $ TVHrsDayChild <int> NA, NA, NA, 4, NA, 5
#> $ CompHrsDayChild <int> NA, NA, NA, 1, NA, 0
#> $ Alcohol12PlusYr <fct> Yes, Yes, Yes, NA, Yes, NA
#> $ AlcoholDay <int> NA, NA, NA, NA, 2, NA
#> $ AlcoholYear <int> 0, 0, 0, NA, 20, NA
#> $ SmokeNow <fct> No, No, No, NA, Yes, NA
#> $ Smoke100 <fct> Yes, Yes, Yes, NA, Yes, NA
#> $ Smoke100n <fct> Smoker, Smoker, Smoker, NA, Smoker, NA
#> $ SmokeAge <int> 18, 18, 18, NA, 38, NA
#> $ Marijuana <fct> Yes, Yes, Yes, NA, Yes, NA
#> $ AgeFirstMarij <int> 17, 17, 17, NA, 18, NA
#> $ RegularMarij <fct> No, No, No, NA, No, NA
#> $ AgeRegMarij <int> NA, NA, NA, NA, NA, NA
#> $ HardDrugs <fct> Yes, Yes, Yes, NA, Yes, NA
#> $ SexEver <fct> Yes, Yes, Yes, NA, Yes, NA
#> $ SexAge <int> 16, 16, 16, NA, 12, NA
#> $ SexNumPartnLife <int> 8, 8, 8, NA, 10, NA
#> $ SexNumPartYear <int> 1, 1, 1, NA, 1, NA
#> $ SameSex <fct> No, No, No, NA, Yes, NA
#> $ SexOrientation <fct> Heterosexual, Heterosexual, Heterosexual, NA, H…
#> $ PregnantNow <fct> NA, NA, NA, NA, NA, NA
# The pipe way of chaining
NHANES %>%
head() %>%
glimpse()
#> Observations: 6
#> Variables: 76
#> $ ID <int> 51624, 51624, 51624, 51625, 51630, 51638
#> $ SurveyYr <fct> 2009_10, 2009_10, 2009_10, 2009_10, 2009_10, 20…
#> $ Gender <fct> male, male, male, male, female, male
#> $ Age <int> 34, 34, 34, 4, 49, 9
#> $ AgeDecade <fct> 30-39, 30-39, 30-39, 0-9, 40-49, 0-9
#> $ AgeMonths <int> 409, 409, 409, 49, 596, 115
#> $ Race1 <fct> White, White, White, Other, White, White
#> $ Race3 <fct> NA, NA, NA, NA, NA, NA
#> $ Education <fct> High School, High School, High School, NA, Some…
#> $ MaritalStatus <fct> Married, Married, Married, NA, LivePartner, NA
#> $ HHIncome <fct> 25000-34999, 25000-34999, 25000-34999, 20000-24…
#> $ HHIncomeMid <int> 30000, 30000, 30000, 22500, 40000, 87500
#> $ Poverty <dbl> 1.36, 1.36, 1.36, 1.07, 1.91, 1.84
#> $ HomeRooms <int> 6, 6, 6, 9, 5, 6
#> $ HomeOwn <fct> Own, Own, Own, Own, Rent, Rent
#> $ Work <fct> NotWorking, NotWorking, NotWorking, NA, NotWork…
#> $ Weight <dbl> 87.4, 87.4, 87.4, 17.0, 86.7, 29.8
#> $ Length <dbl> NA, NA, NA, NA, NA, NA
#> $ HeadCirc <dbl> NA, NA, NA, NA, NA, NA
#> $ Height <dbl> 164.7, 164.7, 164.7, 105.4, 168.4, 133.1
#> $ BMI <dbl> 32.22, 32.22, 32.22, 15.30, 30.57, 16.82
#> $ BMICatUnder20yrs <fct> NA, NA, NA, NA, NA, NA
#> $ BMI_WHO <fct> 30.0_plus, 30.0_plus, 30.0_plus, 12.0_18.5, 30.…
#> $ Pulse <int> 70, 70, 70, NA, 86, 82
#> $ BPSysAve <int> 113, 113, 113, NA, 112, 86
#> $ BPDiaAve <int> 85, 85, 85, NA, 75, 47
#> $ BPSys1 <int> 114, 114, 114, NA, 118, 84
#> $ BPDia1 <int> 88, 88, 88, NA, 82, 50
#> $ BPSys2 <int> 114, 114, 114, NA, 108, 84
#> $ BPDia2 <int> 88, 88, 88, NA, 74, 50
#> $ BPSys3 <int> 112, 112, 112, NA, 116, 88
#> $ BPDia3 <int> 82, 82, 82, NA, 76, 44
#> $ Testosterone <dbl> NA, NA, NA, NA, NA, NA
#> $ DirectChol <dbl> 1.29, 1.29, 1.29, NA, 1.16, 1.34
#> $ TotChol <dbl> 3.49, 3.49, 3.49, NA, 6.70, 4.86
#> $ UrineVol1 <int> 352, 352, 352, NA, 77, 123
#> $ UrineFlow1 <dbl> NA, NA, NA, NA, 0.094, 1.538
#> $ UrineVol2 <int> NA, NA, NA, NA, NA, NA
#> $ UrineFlow2 <dbl> NA, NA, NA, NA, NA, NA
#> $ Diabetes <fct> No, No, No, No, No, No
#> $ DiabetesAge <int> NA, NA, NA, NA, NA, NA
#> $ HealthGen <fct> Good, Good, Good, NA, Good, NA
#> $ DaysPhysHlthBad <int> 0, 0, 0, NA, 0, NA
#> $ DaysMentHlthBad <int> 15, 15, 15, NA, 10, NA
#> $ LittleInterest <fct> Most, Most, Most, NA, Several, NA
#> $ Depressed <fct> Several, Several, Several, NA, Several, NA
#> $ nPregnancies <int> NA, NA, NA, NA, 2, NA
#> $ nBabies <int> NA, NA, NA, NA, 2, NA
#> $ Age1stBaby <int> NA, NA, NA, NA, 27, NA
#> $ SleepHrsNight <int> 4, 4, 4, NA, 8, NA
#> $ SleepTrouble <fct> Yes, Yes, Yes, NA, Yes, NA
#> $ PhysActive <fct> No, No, No, NA, No, NA
#> $ PhysActiveDays <int> NA, NA, NA, NA, NA, NA
#> $ TVHrsDay <fct> NA, NA, NA, NA, NA, NA
#> $ CompHrsDay <fct> NA, NA, NA, NA, NA, NA
#> $ TVHrsDayChild <int> NA, NA, NA, 4, NA, 5
#> $ CompHrsDayChild <int> NA, NA, NA, 1, NA, 0
#> $ Alcohol12PlusYr <fct> Yes, Yes, Yes, NA, Yes, NA
#> $ AlcoholDay <int> NA, NA, NA, NA, 2, NA
#> $ AlcoholYear <int> 0, 0, 0, NA, 20, NA
#> $ SmokeNow <fct> No, No, No, NA, Yes, NA
#> $ Smoke100 <fct> Yes, Yes, Yes, NA, Yes, NA
#> $ Smoke100n <fct> Smoker, Smoker, Smoker, NA, Smoker, NA
#> $ SmokeAge <int> 18, 18, 18, NA, 38, NA
#> $ Marijuana <fct> Yes, Yes, Yes, NA, Yes, NA
#> $ AgeFirstMarij <int> 17, 17, 17, NA, 18, NA
#> $ RegularMarij <fct> No, No, No, NA, No, NA
#> $ AgeRegMarij <int> NA, NA, NA, NA, NA, NA
#> $ HardDrugs <fct> Yes, Yes, Yes, NA, Yes, NA
#> $ SexEver <fct> Yes, Yes, Yes, NA, Yes, NA
#> $ SexAge <int> 16, 16, 16, NA, 12, NA
#> $ SexNumPartnLife <int> 8, 8, 8, NA, 10, NA
#> $ SexNumPartYear <int> 1, 1, 1, NA, 1, NA
#> $ SameSex <fct> No, No, No, NA, Yes, NA
#> $ SexOrientation <fct> Heterosexual, Heterosexual, Heterosexual, NA, H…
#> $ PregnantNow <fct> NA, NA, NA, NA, NA, NA
mutate(): Transforming or adding variables
When you need to add a new column or modify an existing one, you can use the
mutate() function. Get it, mutate as in to change 😉
# Modify an existing variable
NHANES %>%
mutate(Height = Height / 100)
#> # A tibble: 10,000 x 76
#> ID SurveyYr Gender Age AgeDecade AgeMonths Race1 Race3 Education
#> <int> <fct> <fct> <int> <fct> <int> <fct> <fct> <fct>
#> 1 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 2 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 3 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 4 51625 2009_10 male 4 " 0-9" 49 Other <NA> <NA>
#> 5 51630 2009_10 female 49 " 40-49" 596 White <NA> Some Col…
#> 6 51638 2009_10 male 9 " 0-9" 115 White <NA> <NA>
#> 7 51646 2009_10 male 8 " 0-9" 101 White <NA> <NA>
#> 8 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 9 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 10 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> # … with 9,990 more rows, and 67 more variables: MaritalStatus <fct>,
#> # HHIncome <fct>, HHIncomeMid <int>, Poverty <dbl>, HomeRooms <int>,
#> # HomeOwn <fct>, Work <fct>, Weight <dbl>, Length <dbl>, HeadCirc <dbl>,
#> # Height <dbl>, BMI <dbl>, BMICatUnder20yrs <fct>, BMI_WHO <fct>,
#> # Pulse <int>, BPSysAve <int>, BPDiaAve <int>, BPSys1 <int>,
#> # BPDia1 <int>, BPSys2 <int>, BPDia2 <int>, BPSys3 <int>, BPDia3 <int>,
#> # Testosterone <dbl>, DirectChol <dbl>, TotChol <dbl>, UrineVol1 <int>,
#> # UrineFlow1 <dbl>, UrineVol2 <int>, UrineFlow2 <dbl>, Diabetes <fct>,
#> # DiabetesAge <int>, HealthGen <fct>, DaysPhysHlthBad <int>,
#> # DaysMentHlthBad <int>, LittleInterest <fct>, Depressed <fct>,
#> # nPregnancies <int>, nBabies <int>, Age1stBaby <int>,
#> # SleepHrsNight <int>, SleepTrouble <fct>, PhysActive <fct>,
#> # PhysActiveDays <int>, TVHrsDay <fct>, CompHrsDay <fct>,
#> # TVHrsDayChild <int>, CompHrsDayChild <int>, Alcohol12PlusYr <fct>,
#> # AlcoholDay <int>, AlcoholYear <int>, SmokeNow <fct>, Smoke100 <fct>,
#> # Smoke100n <fct>, SmokeAge <int>, Marijuana <fct>, AgeFirstMarij <int>,
#> # RegularMarij <fct>, AgeRegMarij <int>, HardDrugs <fct>, SexEver <fct>,
#> # SexAge <int>, SexNumPartnLife <int>, SexNumPartYear <int>,
#> # SameSex <fct>, SexOrientation <fct>, PregnantNow <fct>
# Or create a new variable based on a condition
NHANES %>%
mutate(HighlyActive = if_else(PhysActiveDays >= 5, "yes", "no"))
#> # A tibble: 10,000 x 77
#> ID SurveyYr Gender Age AgeDecade AgeMonths Race1 Race3 Education
#> <int> <fct> <fct> <int> <fct> <int> <fct> <fct> <fct>
#> 1 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 2 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 3 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 4 51625 2009_10 male 4 " 0-9" 49 Other <NA> <NA>
#> 5 51630 2009_10 female 49 " 40-49" 596 White <NA> Some Col…
#> 6 51638 2009_10 male 9 " 0-9" 115 White <NA> <NA>
#> 7 51646 2009_10 male 8 " 0-9" 101 White <NA> <NA>
#> 8 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 9 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 10 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> # … with 9,990 more rows, and 68 more variables: MaritalStatus <fct>,
#> # HHIncome <fct>, HHIncomeMid <int>, Poverty <dbl>, HomeRooms <int>,
#> # HomeOwn <fct>, Work <fct>, Weight <dbl>, Length <dbl>, HeadCirc <dbl>,
#> # Height <dbl>, BMI <dbl>, BMICatUnder20yrs <fct>, BMI_WHO <fct>,
#> # Pulse <int>, BPSysAve <int>, BPDiaAve <int>, BPSys1 <int>,
#> # BPDia1 <int>, BPSys2 <int>, BPDia2 <int>, BPSys3 <int>, BPDia3 <int>,
#> # Testosterone <dbl>, DirectChol <dbl>, TotChol <dbl>, UrineVol1 <int>,
#> # UrineFlow1 <dbl>, UrineVol2 <int>, UrineFlow2 <dbl>, Diabetes <fct>,
#> # DiabetesAge <int>, HealthGen <fct>, DaysPhysHlthBad <int>,
#> # DaysMentHlthBad <int>, LittleInterest <fct>, Depressed <fct>,
#> # nPregnancies <int>, nBabies <int>, Age1stBaby <int>,
#> # SleepHrsNight <int>, SleepTrouble <fct>, PhysActive <fct>,
#> # PhysActiveDays <int>, TVHrsDay <fct>, CompHrsDay <fct>,
#> # TVHrsDayChild <int>, CompHrsDayChild <int>, Alcohol12PlusYr <fct>,
#> # AlcoholDay <int>, AlcoholYear <int>, SmokeNow <fct>, Smoke100 <fct>,
#> # Smoke100n <fct>, SmokeAge <int>, Marijuana <fct>, AgeFirstMarij <int>,
#> # RegularMarij <fct>, AgeRegMarij <int>, HardDrugs <fct>, SexEver <fct>,
#> # SexAge <int>, SexNumPartnLife <int>, SexNumPartYear <int>,
#> # SameSex <fct>, SexOrientation <fct>, PregnantNow <fct>,
#> # HighlyActive <chr>
# Create or replace multiple variables by using the ","
NHANES %>%
mutate(new_column = "only one value",
Height = Height / 100)
#> # A tibble: 10,000 x 77
#> ID SurveyYr Gender Age AgeDecade AgeMonths Race1 Race3 Education
#> <int> <fct> <fct> <int> <fct> <int> <fct> <fct> <fct>
#> 1 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 2 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 3 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 4 51625 2009_10 male 4 " 0-9" 49 Other <NA> <NA>
#> 5 51630 2009_10 female 49 " 40-49" 596 White <NA> Some Col…
#> 6 51638 2009_10 male 9 " 0-9" 115 White <NA> <NA>
#> 7 51646 2009_10 male 8 " 0-9" 101 White <NA> <NA>
#> 8 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 9 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 10 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> # … with 9,990 more rows, and 68 more variables: MaritalStatus <fct>,
#> # HHIncome <fct>, HHIncomeMid <int>, Poverty <dbl>, HomeRooms <int>,
#> # HomeOwn <fct>, Work <fct>, Weight <dbl>, Length <dbl>, HeadCirc <dbl>,
#> # Height <dbl>, BMI <dbl>, BMICatUnder20yrs <fct>, BMI_WHO <fct>,
#> # Pulse <int>, BPSysAve <int>, BPDiaAve <int>, BPSys1 <int>,
#> # BPDia1 <int>, BPSys2 <int>, BPDia2 <int>, BPSys3 <int>, BPDia3 <int>,
#> # Testosterone <dbl>, DirectChol <dbl>, TotChol <dbl>, UrineVol1 <int>,
#> # UrineFlow1 <dbl>, UrineVol2 <int>, UrineFlow2 <dbl>, Diabetes <fct>,
#> # DiabetesAge <int>, HealthGen <fct>, DaysPhysHlthBad <int>,
#> # DaysMentHlthBad <int>, LittleInterest <fct>, Depressed <fct>,
#> # nPregnancies <int>, nBabies <int>, Age1stBaby <int>,
#> # SleepHrsNight <int>, SleepTrouble <fct>, PhysActive <fct>,
#> # PhysActiveDays <int>, TVHrsDay <fct>, CompHrsDay <fct>,
#> # TVHrsDayChild <int>, CompHrsDayChild <int>, Alcohol12PlusYr <fct>,
#> # AlcoholDay <int>, AlcoholYear <int>, SmokeNow <fct>, Smoke100 <fct>,
#> # Smoke100n <fct>, SmokeAge <int>, Marijuana <fct>, AgeFirstMarij <int>,
#> # RegularMarij <fct>, AgeRegMarij <int>, HardDrugs <fct>, SexEver <fct>,
#> # SexAge <int>, SexNumPartnLife <int>, SexNumPartYear <int>,
#> # SameSex <fct>, SexOrientation <fct>, PregnantNow <fct>,
#> # new_column <chr>
# Create new variable with existing variables and save the changes
NHANES_update <- NHANES %>%
mutate(UrineVolAverage = (UrineVol1 + UrineVol2) / 2)
Use the table below as a reference for logical conditions you can use in the
if_else function.
| Operator | Description |
|---|---|
| < | less than |
| <= | less than or equal to |
| > | greater than |
| >= | greater than or equal to |
| == | equal to |
| != | not equal to |
| !x | Not x |
| x | y | x OR y |
| x & y | x AND y |
Exercise: Piping, transforming, and adding
Time: 10 min
Here’s a chance to practise. Again, using the exercises-wrangling.R script in
the R folder, take the code below and replace the ___ with the appropriate
variable names. Complete the tasks below. (Suggestion: Create a new “Section”
in the script for this exercise via Ctrl-Shift-R).
- Create a new variable called “UrineVolAverage” by calculating the average urine volumne (from “UrineVol1” and “UrineVol2”).
- Modify/replace the “Pulse” variable to beats per second (currently is beats per minute).
- Create a new variable called “YoungChild” when age is less than 6 years.
# Check the names of the variables
colnames(NHANES)
# Pipe the data into mutate function and:
NHANES_modified <- ___ %>% # dataset
mutate(
# 1. Calculate average urine volume
___ = ___,
# 2. Modify Pulse variable
___ = ___,
# 3. Create YoungChild variable using a condition
___ = if_else(___, TRUE, FALSE)
)
NHANES_modified
Notice something about the “UrineVolAverage” values?
For the “UrineVolAverage” values, they are probably almost entirely Click for the solution
# Check the names of the variables
colnames(NHANES)
# Pipe the data into mutate function and:
NHANES_modified <- NHANES %>% # dataset
mutate(
# 1. Calculate average urine volume
UrineVolAverage = (UrineVol1 + UrineVol2) / 2,
# 2. Modify Pulse variable
Pulse = Pulse / 60,
# 3. Create YoungChild variable using a condition
YoungChild = if_else(Age < 6, TRUE, FALSE)
)
NA (aka
missing). That’s because NA values are infectious. See how “UrineVol2” has mostly
NA values too? When you calculate something that has NA, you get another NA.
This is something to be careful about. So always check your calculations!
select(): Select specific data by the variables
Sometimes you only need certain variables from a dataset, not all of them. Or
maybe you want to remove certain ones or select variables that match a pattern.
This is when you use select().
# Select columns/variables by name, without quotes
NHANES %>%
select(Age, Gender, BMI)
#> # A tibble: 10,000 x 3
#> Age Gender BMI
#> <int> <fct> <dbl>
#> 1 34 male 32.2
#> 2 34 male 32.2
#> 3 34 male 32.2
#> 4 4 male 15.3
#> 5 49 female 30.6
#> 6 9 male 16.8
#> 7 8 male 20.6
#> 8 45 female 27.2
#> 9 45 female 27.2
#> 10 45 female 27.2
#> # … with 9,990 more rows
# To *not* select a variable, us minus (-)
NHANES %>%
select(-HeadCirc)
#> # A tibble: 10,000 x 75
#> ID SurveyYr Gender Age AgeDecade AgeMonths Race1 Race3 Education
#> <int> <fct> <fct> <int> <fct> <int> <fct> <fct> <fct>
#> 1 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 2 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 3 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 4 51625 2009_10 male 4 " 0-9" 49 Other <NA> <NA>
#> 5 51630 2009_10 female 49 " 40-49" 596 White <NA> Some Col…
#> 6 51638 2009_10 male 9 " 0-9" 115 White <NA> <NA>
#> 7 51646 2009_10 male 8 " 0-9" 101 White <NA> <NA>
#> 8 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 9 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 10 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> # … with 9,990 more rows, and 66 more variables: MaritalStatus <fct>,
#> # HHIncome <fct>, HHIncomeMid <int>, Poverty <dbl>, HomeRooms <int>,
#> # HomeOwn <fct>, Work <fct>, Weight <dbl>, Length <dbl>, Height <dbl>,
#> # BMI <dbl>, BMICatUnder20yrs <fct>, BMI_WHO <fct>, Pulse <int>,
#> # BPSysAve <int>, BPDiaAve <int>, BPSys1 <int>, BPDia1 <int>,
#> # BPSys2 <int>, BPDia2 <int>, BPSys3 <int>, BPDia3 <int>,
#> # Testosterone <dbl>, DirectChol <dbl>, TotChol <dbl>, UrineVol1 <int>,
#> # UrineFlow1 <dbl>, UrineVol2 <int>, UrineFlow2 <dbl>, Diabetes <fct>,
#> # DiabetesAge <int>, HealthGen <fct>, DaysPhysHlthBad <int>,
#> # DaysMentHlthBad <int>, LittleInterest <fct>, Depressed <fct>,
#> # nPregnancies <int>, nBabies <int>, Age1stBaby <int>,
#> # SleepHrsNight <int>, SleepTrouble <fct>, PhysActive <fct>,
#> # PhysActiveDays <int>, TVHrsDay <fct>, CompHrsDay <fct>,
#> # TVHrsDayChild <int>, CompHrsDayChild <int>, Alcohol12PlusYr <fct>,
#> # AlcoholDay <int>, AlcoholYear <int>, SmokeNow <fct>, Smoke100 <fct>,
#> # Smoke100n <fct>, SmokeAge <int>, Marijuana <fct>, AgeFirstMarij <int>,
#> # RegularMarij <fct>, AgeRegMarij <int>, HardDrugs <fct>, SexEver <fct>,
#> # SexAge <int>, SexNumPartnLife <int>, SexNumPartYear <int>,
#> # SameSex <fct>, SexOrientation <fct>, PregnantNow <fct>
# When you have many variables with similar names, use "matching" functions
NHANES %>%
select(starts_with("BP"), contains("Vol"))
#> # A tibble: 10,000 x 10
#> BPSysAve BPDiaAve BPSys1 BPDia1 BPSys2 BPDia2 BPSys3 BPDia3 UrineVol1
#> <int> <int> <int> <int> <int> <int> <int> <int> <int>
#> 1 113 85 114 88 114 88 112 82 352
#> 2 113 85 114 88 114 88 112 82 352
#> 3 113 85 114 88 114 88 112 82 352
#> 4 NA NA NA NA NA NA NA NA NA
#> 5 112 75 118 82 108 74 116 76 77
#> 6 86 47 84 50 84 50 88 44 123
#> 7 107 37 114 46 108 36 106 38 238
#> 8 118 64 106 62 118 68 118 60 106
#> 9 118 64 106 62 118 68 118 60 106
#> 10 118 64 106 62 118 68 118 60 106
#> # … with 9,990 more rows, and 1 more variable: UrineVol2 <int>
For more information on using the pattern functions such as starts_with(),
check ?select_helpers. If you are familiar with regular expressions, you can
use matches().
rename(): Rename specific columns
If you need to rename a column, that’s super easy! Use the rename() function!
# rename using the form "newname = oldname"
NHANES %>%
rename(NumberBabies = nBabies)
#> # A tibble: 10,000 x 76
#> ID SurveyYr Gender Age AgeDecade AgeMonths Race1 Race3 Education
#> <int> <fct> <fct> <int> <fct> <int> <fct> <fct> <fct>
#> 1 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 2 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 3 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 4 51625 2009_10 male 4 " 0-9" 49 Other <NA> <NA>
#> 5 51630 2009_10 female 49 " 40-49" 596 White <NA> Some Col…
#> 6 51638 2009_10 male 9 " 0-9" 115 White <NA> <NA>
#> 7 51646 2009_10 male 8 " 0-9" 101 White <NA> <NA>
#> 8 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 9 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 10 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> # … with 9,990 more rows, and 67 more variables: MaritalStatus <fct>,
#> # HHIncome <fct>, HHIncomeMid <int>, Poverty <dbl>, HomeRooms <int>,
#> # HomeOwn <fct>, Work <fct>, Weight <dbl>, Length <dbl>, HeadCirc <dbl>,
#> # Height <dbl>, BMI <dbl>, BMICatUnder20yrs <fct>, BMI_WHO <fct>,
#> # Pulse <int>, BPSysAve <int>, BPDiaAve <int>, BPSys1 <int>,
#> # BPDia1 <int>, BPSys2 <int>, BPDia2 <int>, BPSys3 <int>, BPDia3 <int>,
#> # Testosterone <dbl>, DirectChol <dbl>, TotChol <dbl>, UrineVol1 <int>,
#> # UrineFlow1 <dbl>, UrineVol2 <int>, UrineFlow2 <dbl>, Diabetes <fct>,
#> # DiabetesAge <int>, HealthGen <fct>, DaysPhysHlthBad <int>,
#> # DaysMentHlthBad <int>, LittleInterest <fct>, Depressed <fct>,
#> # nPregnancies <int>, NumberBabies <int>, Age1stBaby <int>,
#> # SleepHrsNight <int>, SleepTrouble <fct>, PhysActive <fct>,
#> # PhysActiveDays <int>, TVHrsDay <fct>, CompHrsDay <fct>,
#> # TVHrsDayChild <int>, CompHrsDayChild <int>, Alcohol12PlusYr <fct>,
#> # AlcoholDay <int>, AlcoholYear <int>, SmokeNow <fct>, Smoke100 <fct>,
#> # Smoke100n <fct>, SmokeAge <int>, Marijuana <fct>, AgeFirstMarij <int>,
#> # RegularMarij <fct>, AgeRegMarij <int>, HardDrugs <fct>, SexEver <fct>,
#> # SexAge <int>, SexNumPartnLife <int>, SexNumPartYear <int>,
#> # SameSex <fct>, SexOrientation <fct>, PregnantNow <fct>
filter(): Filtering/subsetting the data by row
Subsetting/filtering data is a very common activity in data analysis, for example,
keeping only those older than 30 years. Using the filter() function lets you
do this. Filtering uses logic for conditions, so refer to the table above for the
logic operators.
A warning: Filtering using logic. Humans are really bad at logic. Make sure that you are very certain that what you think your logic is doing is what the code is actually doing. Confirm the findings! Lots of mistakes can be made at this stage, especially with complex logic conditions and filtering.
# when gender is equal to
NHANES %>%
filter(Gender == "female")
#> # A tibble: 5,020 x 76
#> ID SurveyYr Gender Age AgeDecade AgeMonths Race1 Race3 Education
#> <int> <fct> <fct> <int> <fct> <int> <fct> <fct> <fct>
#> 1 51630 2009_10 female 49 " 40-49" 596 White <NA> Some Col…
#> 2 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 3 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 4 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 5 51659 2009_10 female 10 " 10-19" 123 White <NA> <NA>
#> 6 51666 2009_10 female 58 " 50-59" 700 Mexi… <NA> High Sch…
#> 7 51671 2009_10 female 9 " 0-9" 112 Black <NA> <NA>
#> 8 51685 2009_10 female 56 " 50-59" 677 White <NA> College …
#> 9 51685 2009_10 female 56 " 50-59" 677 White <NA> College …
#> 10 51691 2009_10 female 57 " 50-59" 694 White <NA> High Sch…
#> # … with 5,010 more rows, and 67 more variables: MaritalStatus <fct>,
#> # HHIncome <fct>, HHIncomeMid <int>, Poverty <dbl>, HomeRooms <int>,
#> # HomeOwn <fct>, Work <fct>, Weight <dbl>, Length <dbl>, HeadCirc <dbl>,
#> # Height <dbl>, BMI <dbl>, BMICatUnder20yrs <fct>, BMI_WHO <fct>,
#> # Pulse <int>, BPSysAve <int>, BPDiaAve <int>, BPSys1 <int>,
#> # BPDia1 <int>, BPSys2 <int>, BPDia2 <int>, BPSys3 <int>, BPDia3 <int>,
#> # Testosterone <dbl>, DirectChol <dbl>, TotChol <dbl>, UrineVol1 <int>,
#> # UrineFlow1 <dbl>, UrineVol2 <int>, UrineFlow2 <dbl>, Diabetes <fct>,
#> # DiabetesAge <int>, HealthGen <fct>, DaysPhysHlthBad <int>,
#> # DaysMentHlthBad <int>, LittleInterest <fct>, Depressed <fct>,
#> # nPregnancies <int>, nBabies <int>, Age1stBaby <int>,
#> # SleepHrsNight <int>, SleepTrouble <fct>, PhysActive <fct>,
#> # PhysActiveDays <int>, TVHrsDay <fct>, CompHrsDay <fct>,
#> # TVHrsDayChild <int>, CompHrsDayChild <int>, Alcohol12PlusYr <fct>,
#> # AlcoholDay <int>, AlcoholYear <int>, SmokeNow <fct>, Smoke100 <fct>,
#> # Smoke100n <fct>, SmokeAge <int>, Marijuana <fct>, AgeFirstMarij <int>,
#> # RegularMarij <fct>, AgeRegMarij <int>, HardDrugs <fct>, SexEver <fct>,
#> # SexAge <int>, SexNumPartnLife <int>, SexNumPartYear <int>,
#> # SameSex <fct>, SexOrientation <fct>, PregnantNow <fct>
# when gender is *not* equal to
NHANES %>%
filter(Gender != "female")
#> # A tibble: 4,980 x 76
#> ID SurveyYr Gender Age AgeDecade AgeMonths Race1 Race3 Education
#> <int> <fct> <fct> <int> <fct> <int> <fct> <fct> <fct>
#> 1 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 2 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 3 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 4 51625 2009_10 male 4 " 0-9" 49 Other <NA> <NA>
#> 5 51638 2009_10 male 9 " 0-9" 115 White <NA> <NA>
#> 6 51646 2009_10 male 8 " 0-9" 101 White <NA> <NA>
#> 7 51654 2009_10 male 66 " 60-69" 795 White <NA> Some Col…
#> 8 51656 2009_10 male 58 " 50-59" 707 White <NA> College …
#> 9 51657 2009_10 male 54 " 50-59" 654 White <NA> 9 - 11th…
#> 10 51667 2009_10 male 50 " 50-59" 603 White <NA> Some Col…
#> # … with 4,970 more rows, and 67 more variables: MaritalStatus <fct>,
#> # HHIncome <fct>, HHIncomeMid <int>, Poverty <dbl>, HomeRooms <int>,
#> # HomeOwn <fct>, Work <fct>, Weight <dbl>, Length <dbl>, HeadCirc <dbl>,
#> # Height <dbl>, BMI <dbl>, BMICatUnder20yrs <fct>, BMI_WHO <fct>,
#> # Pulse <int>, BPSysAve <int>, BPDiaAve <int>, BPSys1 <int>,
#> # BPDia1 <int>, BPSys2 <int>, BPDia2 <int>, BPSys3 <int>, BPDia3 <int>,
#> # Testosterone <dbl>, DirectChol <dbl>, TotChol <dbl>, UrineVol1 <int>,
#> # UrineFlow1 <dbl>, UrineVol2 <int>, UrineFlow2 <dbl>, Diabetes <fct>,
#> # DiabetesAge <int>, HealthGen <fct>, DaysPhysHlthBad <int>,
#> # DaysMentHlthBad <int>, LittleInterest <fct>, Depressed <fct>,
#> # nPregnancies <int>, nBabies <int>, Age1stBaby <int>,
#> # SleepHrsNight <int>, SleepTrouble <fct>, PhysActive <fct>,
#> # PhysActiveDays <int>, TVHrsDay <fct>, CompHrsDay <fct>,
#> # TVHrsDayChild <int>, CompHrsDayChild <int>, Alcohol12PlusYr <fct>,
#> # AlcoholDay <int>, AlcoholYear <int>, SmokeNow <fct>, Smoke100 <fct>,
#> # Smoke100n <fct>, SmokeAge <int>, Marijuana <fct>, AgeFirstMarij <int>,
#> # RegularMarij <fct>, AgeRegMarij <int>, HardDrugs <fct>, SexEver <fct>,
#> # SexAge <int>, SexNumPartnLife <int>, SexNumPartYear <int>,
#> # SameSex <fct>, SexOrientation <fct>, PregnantNow <fct>
# when BMI is equal to
NHANES %>%
filter(BMI == 25)
#> # A tibble: 35 x 76
#> ID SurveyYr Gender Age AgeDecade AgeMonths Race1 Race3 Education
#> <int> <fct> <fct> <int> <fct> <int> <fct> <fct> <fct>
#> 1 57116 2009_10 male 24 " 20-29" 288 Other <NA> 8th Grade
#> 2 57116 2009_10 male 24 " 20-29" 288 Other <NA> 8th Grade
#> 3 62222 2011_12 male 32 " 30-39" NA White White College …
#> 4 62222 2011_12 male 32 " 30-39" NA White White College …
#> 5 62222 2011_12 male 32 " 30-39" NA White White College …
#> 6 62222 2011_12 male 32 " 30-39" NA White White College …
#> 7 62277 2011_12 female 55 " 50-59" NA White White Some Col…
#> 8 62578 2011_12 female 6 " 0-9" NA White White <NA>
#> 9 62806 2011_12 male 53 " 50-59" NA Black Black 9 - 11th…
#> 10 62881 2011_12 female 57 " 50-59" NA White White College …
#> # … with 25 more rows, and 67 more variables: MaritalStatus <fct>,
#> # HHIncome <fct>, HHIncomeMid <int>, Poverty <dbl>, HomeRooms <int>,
#> # HomeOwn <fct>, Work <fct>, Weight <dbl>, Length <dbl>, HeadCirc <dbl>,
#> # Height <dbl>, BMI <dbl>, BMICatUnder20yrs <fct>, BMI_WHO <fct>,
#> # Pulse <int>, BPSysAve <int>, BPDiaAve <int>, BPSys1 <int>,
#> # BPDia1 <int>, BPSys2 <int>, BPDia2 <int>, BPSys3 <int>, BPDia3 <int>,
#> # Testosterone <dbl>, DirectChol <dbl>, TotChol <dbl>, UrineVol1 <int>,
#> # UrineFlow1 <dbl>, UrineVol2 <int>, UrineFlow2 <dbl>, Diabetes <fct>,
#> # DiabetesAge <int>, HealthGen <fct>, DaysPhysHlthBad <int>,
#> # DaysMentHlthBad <int>, LittleInterest <fct>, Depressed <fct>,
#> # nPregnancies <int>, nBabies <int>, Age1stBaby <int>,
#> # SleepHrsNight <int>, SleepTrouble <fct>, PhysActive <fct>,
#> # PhysActiveDays <int>, TVHrsDay <fct>, CompHrsDay <fct>,
#> # TVHrsDayChild <int>, CompHrsDayChild <int>, Alcohol12PlusYr <fct>,
#> # AlcoholDay <int>, AlcoholYear <int>, SmokeNow <fct>, Smoke100 <fct>,
#> # Smoke100n <fct>, SmokeAge <int>, Marijuana <fct>, AgeFirstMarij <int>,
#> # RegularMarij <fct>, AgeRegMarij <int>, HardDrugs <fct>, SexEver <fct>,
#> # SexAge <int>, SexNumPartnLife <int>, SexNumPartYear <int>,
#> # SameSex <fct>, SexOrientation <fct>, PregnantNow <fct>
# when BMI is equal to or more than
NHANES %>%
filter(BMI >= 25)
#> # A tibble: 5,422 x 76
#> ID SurveyYr Gender Age AgeDecade AgeMonths Race1 Race3 Education
#> <int> <fct> <fct> <int> <fct> <int> <fct> <fct> <fct>
#> 1 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 2 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 3 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 4 51630 2009_10 female 49 " 40-49" 596 White <NA> Some Col…
#> 5 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 6 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 7 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 8 51657 2009_10 male 54 " 50-59" 654 White <NA> 9 - 11th…
#> 9 51666 2009_10 female 58 " 50-59" 700 Mexi… <NA> High Sch…
#> 10 51667 2009_10 male 50 " 50-59" 603 White <NA> Some Col…
#> # … with 5,412 more rows, and 67 more variables: MaritalStatus <fct>,
#> # HHIncome <fct>, HHIncomeMid <int>, Poverty <dbl>, HomeRooms <int>,
#> # HomeOwn <fct>, Work <fct>, Weight <dbl>, Length <dbl>, HeadCirc <dbl>,
#> # Height <dbl>, BMI <dbl>, BMICatUnder20yrs <fct>, BMI_WHO <fct>,
#> # Pulse <int>, BPSysAve <int>, BPDiaAve <int>, BPSys1 <int>,
#> # BPDia1 <int>, BPSys2 <int>, BPDia2 <int>, BPSys3 <int>, BPDia3 <int>,
#> # Testosterone <dbl>, DirectChol <dbl>, TotChol <dbl>, UrineVol1 <int>,
#> # UrineFlow1 <dbl>, UrineVol2 <int>, UrineFlow2 <dbl>, Diabetes <fct>,
#> # DiabetesAge <int>, HealthGen <fct>, DaysPhysHlthBad <int>,
#> # DaysMentHlthBad <int>, LittleInterest <fct>, Depressed <fct>,
#> # nPregnancies <int>, nBabies <int>, Age1stBaby <int>,
#> # SleepHrsNight <int>, SleepTrouble <fct>, PhysActive <fct>,
#> # PhysActiveDays <int>, TVHrsDay <fct>, CompHrsDay <fct>,
#> # TVHrsDayChild <int>, CompHrsDayChild <int>, Alcohol12PlusYr <fct>,
#> # AlcoholDay <int>, AlcoholYear <int>, SmokeNow <fct>, Smoke100 <fct>,
#> # Smoke100n <fct>, SmokeAge <int>, Marijuana <fct>, AgeFirstMarij <int>,
#> # RegularMarij <fct>, AgeRegMarij <int>, HardDrugs <fct>, SexEver <fct>,
#> # SexAge <int>, SexNumPartnLife <int>, SexNumPartYear <int>,
#> # SameSex <fct>, SexOrientation <fct>, PregnantNow <fct>
# when BMI is 25 *and* Gender is female
NHANES %>%
filter(BMI == 25 & Gender == "female")
#> # A tibble: 21 x 76
#> ID SurveyYr Gender Age AgeDecade AgeMonths Race1 Race3 Education
#> <int> <fct> <fct> <int> <fct> <int> <fct> <fct> <fct>
#> 1 62277 2011_12 female 55 " 50-59" NA White White Some Col…
#> 2 62578 2011_12 female 6 " 0-9" NA White White <NA>
#> 3 62881 2011_12 female 57 " 50-59" NA White White College …
#> 4 62881 2011_12 female 57 " 50-59" NA White White College …
#> 5 62881 2011_12 female 57 " 50-59" NA White White College …
#> 6 65572 2011_12 female 79 " 70+" NA Other Asian College …
#> 7 65688 2011_12 female 54 " 50-59" NA White White Some Col…
#> 8 65688 2011_12 female 54 " 50-59" NA White White Some Col…
#> 9 65688 2011_12 female 54 " 50-59" NA White White Some Col…
#> 10 65688 2011_12 female 54 " 50-59" NA White White Some Col…
#> # … with 11 more rows, and 67 more variables: MaritalStatus <fct>,
#> # HHIncome <fct>, HHIncomeMid <int>, Poverty <dbl>, HomeRooms <int>,
#> # HomeOwn <fct>, Work <fct>, Weight <dbl>, Length <dbl>, HeadCirc <dbl>,
#> # Height <dbl>, BMI <dbl>, BMICatUnder20yrs <fct>, BMI_WHO <fct>,
#> # Pulse <int>, BPSysAve <int>, BPDiaAve <int>, BPSys1 <int>,
#> # BPDia1 <int>, BPSys2 <int>, BPDia2 <int>, BPSys3 <int>, BPDia3 <int>,
#> # Testosterone <dbl>, DirectChol <dbl>, TotChol <dbl>, UrineVol1 <int>,
#> # UrineFlow1 <dbl>, UrineVol2 <int>, UrineFlow2 <dbl>, Diabetes <fct>,
#> # DiabetesAge <int>, HealthGen <fct>, DaysPhysHlthBad <int>,
#> # DaysMentHlthBad <int>, LittleInterest <fct>, Depressed <fct>,
#> # nPregnancies <int>, nBabies <int>, Age1stBaby <int>,
#> # SleepHrsNight <int>, SleepTrouble <fct>, PhysActive <fct>,
#> # PhysActiveDays <int>, TVHrsDay <fct>, CompHrsDay <fct>,
#> # TVHrsDayChild <int>, CompHrsDayChild <int>, Alcohol12PlusYr <fct>,
#> # AlcoholDay <int>, AlcoholYear <int>, SmokeNow <fct>, Smoke100 <fct>,
#> # Smoke100n <fct>, SmokeAge <int>, Marijuana <fct>, AgeFirstMarij <int>,
#> # RegularMarij <fct>, AgeRegMarij <int>, HardDrugs <fct>, SexEver <fct>,
#> # SexAge <int>, SexNumPartnLife <int>, SexNumPartYear <int>,
#> # SameSex <fct>, SexOrientation <fct>, PregnantNow <fct>
# when BMI is 25 *or* gender is female
NHANES %>%
filter(BMI == 25 | Gender == "female")
#> # A tibble: 5,034 x 76
#> ID SurveyYr Gender Age AgeDecade AgeMonths Race1 Race3 Education
#> <int> <fct> <fct> <int> <fct> <int> <fct> <fct> <fct>
#> 1 51630 2009_10 female 49 " 40-49" 596 White <NA> Some Col…
#> 2 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 3 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 4 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 5 51659 2009_10 female 10 " 10-19" 123 White <NA> <NA>
#> 6 51666 2009_10 female 58 " 50-59" 700 Mexi… <NA> High Sch…
#> 7 51671 2009_10 female 9 " 0-9" 112 Black <NA> <NA>
#> 8 51685 2009_10 female 56 " 50-59" 677 White <NA> College …
#> 9 51685 2009_10 female 56 " 50-59" 677 White <NA> College …
#> 10 51691 2009_10 female 57 " 50-59" 694 White <NA> High Sch…
#> # … with 5,024 more rows, and 67 more variables: MaritalStatus <fct>,
#> # HHIncome <fct>, HHIncomeMid <int>, Poverty <dbl>, HomeRooms <int>,
#> # HomeOwn <fct>, Work <fct>, Weight <dbl>, Length <dbl>, HeadCirc <dbl>,
#> # Height <dbl>, BMI <dbl>, BMICatUnder20yrs <fct>, BMI_WHO <fct>,
#> # Pulse <int>, BPSysAve <int>, BPDiaAve <int>, BPSys1 <int>,
#> # BPDia1 <int>, BPSys2 <int>, BPDia2 <int>, BPSys3 <int>, BPDia3 <int>,
#> # Testosterone <dbl>, DirectChol <dbl>, TotChol <dbl>, UrineVol1 <int>,
#> # UrineFlow1 <dbl>, UrineVol2 <int>, UrineFlow2 <dbl>, Diabetes <fct>,
#> # DiabetesAge <int>, HealthGen <fct>, DaysPhysHlthBad <int>,
#> # DaysMentHlthBad <int>, LittleInterest <fct>, Depressed <fct>,
#> # nPregnancies <int>, nBabies <int>, Age1stBaby <int>,
#> # SleepHrsNight <int>, SleepTrouble <fct>, PhysActive <fct>,
#> # PhysActiveDays <int>, TVHrsDay <fct>, CompHrsDay <fct>,
#> # TVHrsDayChild <int>, CompHrsDayChild <int>, Alcohol12PlusYr <fct>,
#> # AlcoholDay <int>, AlcoholYear <int>, SmokeNow <fct>, Smoke100 <fct>,
#> # Smoke100n <fct>, SmokeAge <int>, Marijuana <fct>, AgeFirstMarij <int>,
#> # RegularMarij <fct>, AgeRegMarij <int>, HardDrugs <fct>, SexEver <fct>,
#> # SexAge <int>, SexNumPartnLife <int>, SexNumPartYear <int>,
#> # SameSex <fct>, SexOrientation <fct>, PregnantNow <fct>
arrange(): Sorting/(re)arranging your data by column
Arranging/sorting your rows is fairly simple with dplyr. Just use arrange()!
# ascending order by age
NHANES %>%
arrange(Age) %>%
select(Age)
#> # A tibble: 10,000 x 1
#> Age
#> <int>
#> 1 0
#> 2 0
#> 3 0
#> 4 0
#> 5 0
#> 6 0
#> 7 0
#> 8 0
#> 9 0
#> 10 0
#> # … with 9,990 more rows
# Other arrange (for reference)
# descending order by rate
NHANES %>%
arrange(desc(Age)) %>%
select(Age)
#> # A tibble: 10,000 x 1
#> Age
#> <int>
#> 1 80
#> 2 80
#> 3 80
#> 4 80
#> 5 80
#> 6 80
#> 7 80
#> 8 80
#> 9 80
#> 10 80
#> # … with 9,990 more rows
# ascending order by Age and Gender
NHANES %>%
arrange(Age, Gender) %>%
select(Age, Gender)
#> # A tibble: 10,000 x 2
#> Age Gender
#> <int> <fct>
#> 1 0 female
#> 2 0 female
#> 3 0 female
#> 4 0 female
#> 5 0 female
#> 6 0 female
#> 7 0 female
#> 8 0 female
#> 9 0 female
#> 10 0 female
#> # … with 9,990 more rows
Exercise: Filtering and logic, arranging, and selecting
Time: 10 min
Copy and paste the code below into the script exercises-wrangling.R (in a new
Section, Ctrl-Shift-R). Then start replacing the ___ with the appropriate
code to complete the tasks below:
- Filter so only those with BMI more than 20 and less than 40 and keep only those with diabetes.
- Filter to keep those who are working (“Work”) or those who are renting (“HomeOwn”) and those who do not have diabetes. Select the variables age, gender, work status, home ownership, and diabetes status.
- Using sorting and selecting, find out who has had the most number of babies and how old they are.
# To see values of categorical data
summary(NHANES)
# 1. BMI between 20 and 40 and who have diabetes
NHANES %>%
# format: variable >= number
filter(___ >= ___ & ___ <= ___ & ___ == ___)
# 2. Working or renting, and not diabetes
___ %>%
filter(___ == ___ | ___ == ___ & ___ == ___) %>%
select(___)
# 3. How old is person with most number of children.
___ %>%
___(___) %>%
___(___)
Click for a possible solution
# 1. BMI between 20 and 40 and who have diabetes
NHANES %>%
# format: variable >= number
filter(BMI >= 20 & BMI <= 40 & Diabetes == "Yes")
#> # A tibble: 616 x 76
#> ID SurveyYr Gender Age AgeDecade AgeMonths Race1 Race3 Education
#> <int> <fct> <fct> <int> <fct> <int> <fct> <fct> <fct>
#> 1 51702 2009_10 male 44 " 40-49" 539 White <NA> 9 - 11th…
#> 2 51702 2009_10 male 44 " 40-49" 539 White <NA> 9 - 11th…
#> 3 51707 2009_10 female 64 " 60-69" 771 Other <NA> 8th Grade
#> 4 51711 2009_10 female 59 " 50-59" 718 Other <NA> 8th Grade
#> 5 51711 2009_10 female 59 " 50-59" 718 Other <NA> 8th Grade
#> 6 51748 2009_10 male 56 " 50-59" 682 Mexi… <NA> 8th Grade
#> 7 51752 2009_10 female 78 " 70+" 937 Mexi… <NA> 8th Grade
#> 8 51819 2009_10 male 80 <NA> NA White <NA> 9 - 11th…
#> 9 51828 2009_10 female 66 " 60-69" 800 Black <NA> 9 - 11th…
#> 10 51828 2009_10 female 66 " 60-69" 800 Black <NA> 9 - 11th…
#> # … with 606 more rows, and 67 more variables: MaritalStatus <fct>,
#> # HHIncome <fct>, HHIncomeMid <int>, Poverty <dbl>, HomeRooms <int>,
#> # HomeOwn <fct>, Work <fct>, Weight <dbl>, Length <dbl>, HeadCirc <dbl>,
#> # Height <dbl>, BMI <dbl>, BMICatUnder20yrs <fct>, BMI_WHO <fct>,
#> # Pulse <int>, BPSysAve <int>, BPDiaAve <int>, BPSys1 <int>,
#> # BPDia1 <int>, BPSys2 <int>, BPDia2 <int>, BPSys3 <int>, BPDia3 <int>,
#> # Testosterone <dbl>, DirectChol <dbl>, TotChol <dbl>, UrineVol1 <int>,
#> # UrineFlow1 <dbl>, UrineVol2 <int>, UrineFlow2 <dbl>, Diabetes <fct>,
#> # DiabetesAge <int>, HealthGen <fct>, DaysPhysHlthBad <int>,
#> # DaysMentHlthBad <int>, LittleInterest <fct>, Depressed <fct>,
#> # nPregnancies <int>, nBabies <int>, Age1stBaby <int>,
#> # SleepHrsNight <int>, SleepTrouble <fct>, PhysActive <fct>,
#> # PhysActiveDays <int>, TVHrsDay <fct>, CompHrsDay <fct>,
#> # TVHrsDayChild <int>, CompHrsDayChild <int>, Alcohol12PlusYr <fct>,
#> # AlcoholDay <int>, AlcoholYear <int>, SmokeNow <fct>, Smoke100 <fct>,
#> # Smoke100n <fct>, SmokeAge <int>, Marijuana <fct>, AgeFirstMarij <int>,
#> # RegularMarij <fct>, AgeRegMarij <int>, HardDrugs <fct>, SexEver <fct>,
#> # SexAge <int>, SexNumPartnLife <int>, SexNumPartYear <int>,
#> # SameSex <fct>, SexOrientation <fct>, PregnantNow <fct>
# 2. Working or renting, and not diabetes
NHANES %>%
filter(Work == "Working" | HomeOwn == "Rent" & Diabetes == "Yes") %>%
select(Age, Gender, Work, HomeOwn, Diabetes)
#> # A tibble: 4,751 x 5
#> Age Gender Work HomeOwn Diabetes
#> <int> <fct> <fct> <fct> <fct>
#> 1 45 female Working Own No
#> 2 45 female Working Own No
#> 3 45 female Working Own No
#> 4 58 male Working Rent No
#> 5 54 male Working Rent No
#> 6 33 male Working Own No
#> 7 60 male Working Own No
#> 8 57 female Working Own No
#> 9 38 male Working Own No
#> 10 44 male Working Rent Yes
#> # … with 4,741 more rows
# 3. How old is person with most number of children.
NHANES %>%
arrange(desc(nBabies)) %>%
select(Age, nBabies)
#> # A tibble: 10,000 x 2
#> Age nBabies
#> <int> <int>
#> 1 72 12
#> 2 80 11
#> 3 46 11
#> 4 80 9
#> 5 77 9
#> 6 77 9
#> 7 50 8
#> 8 63 8
#> 9 66 8
#> 10 65 8
#> # … with 9,990 more rows
group_by(), summarise(): Create a summary of the data, alone or by a group(s)
Using group_by() on its own does nothing. But, combine it with other functions
like summarise() and you can do some very powerful data processing. This
particular combination of commands allows for the split-apply-combine
technique to be easily used. Many data analysis tasks can be approached using
this paradigm: split the data into groups, apply some analysis to each group,
and then combine the results together. Using group_by() splits the data up and
summarise() then applies an analysis and then combines it back together. The
arguments to group_by() are the column names that contain the categorical
variables for what you want to calculate the summary statistics.
Let’s first explore using summarise() on it’s own, which outputs a single value
(e.g. a max or mean). Like mutate(), you can add multiple “summaries” by adding
new variables separated by commas.
NHANES %>%
summarise(MaxAge = max(Age, na.rm = TRUE),
MeanBMI = mean(BMI, na.rm = TRUE))
#> # A tibble: 1 x 2
#> MaxAge MeanBMI
#> <int> <dbl>
#> 1 80 26.7
Combine it with group_by().
# Grouped by gender
NHANES %>%
group_by(Gender) %>%
summarise(MaxAge = max(Age, na.rm = TRUE),
MeanBMI = mean(BMI, na.rm = TRUE))
#> # A tibble: 2 x 3
#> Gender MaxAge MeanBMI
#> <fct> <int> <dbl>
#> 1 female 80 26.8
#> 2 male 80 26.5
# Grouped by gender and diabetes
NHANES %>%
group_by(Gender, Diabetes) %>%
summarise(MeanAge = mean(Age, na.rm = TRUE),
MeanBMI = mean(BMI, na.rm = TRUE))
#> Warning: Factor `Diabetes` contains implicit NA, consider using
#> `forcats::fct_explicit_na`
#> # A tibble: 6 x 4
#> # Groups: Gender [2]
#> Gender Diabetes MeanAge MeanBMI
#> <fct> <fct> <dbl> <dbl>
#> 1 female No 36.5 26.2
#> 2 female Yes 59.9 33.7
#> 3 female <NA> 1.89 40.8
#> 4 male No 34.3 26.1
#> 5 male Yes 58.6 31.5
#> 6 male <NA> 0.507 22.2
gather(): Converting from wide to long form
The tidyr package is a companion package to dplyr and allows for a wide range of
manipulations of the data structure data. For example, sometimes we have data
in the wide form as it is often easier to enter data in wide form. Wide form can
be useful when presenting as tables. But there are usually problems with wide
form data, especially when it comes to analysing it. Look at the table below,
which is in wide form. Wide form data also tends to be a bit “messier” then long
form. For instance, what do the values in the 1999 column mean? Unless you had
a data dictionary, you would not know what those values mean.
| country | 1999 | 2000 |
|---|---|---|
| Afghanistan | 19987071 | 20595360 |
| Brazil | 172006362 | 174504898 |
| China | 1272915272 | 1280428583 |
The long form on the other hand is usually better suited for analyses and visualizing, especially when doing split-apply-combine techniques. Long form data also tends to be more tidy compared to wide form.
| country | year | population |
|---|---|---|
| Afghanistan | 1999 | 19987071 |
| Afghanistan | 2000 | 20595360 |
| Brazil | 1999 | 172006362 |
| Brazil | 2000 | 174504898 |
| China | 1999 | 1272915272 |
| China | 2000 | 1280428583 |
We can get to this from the wide form through wrangling the data like so:
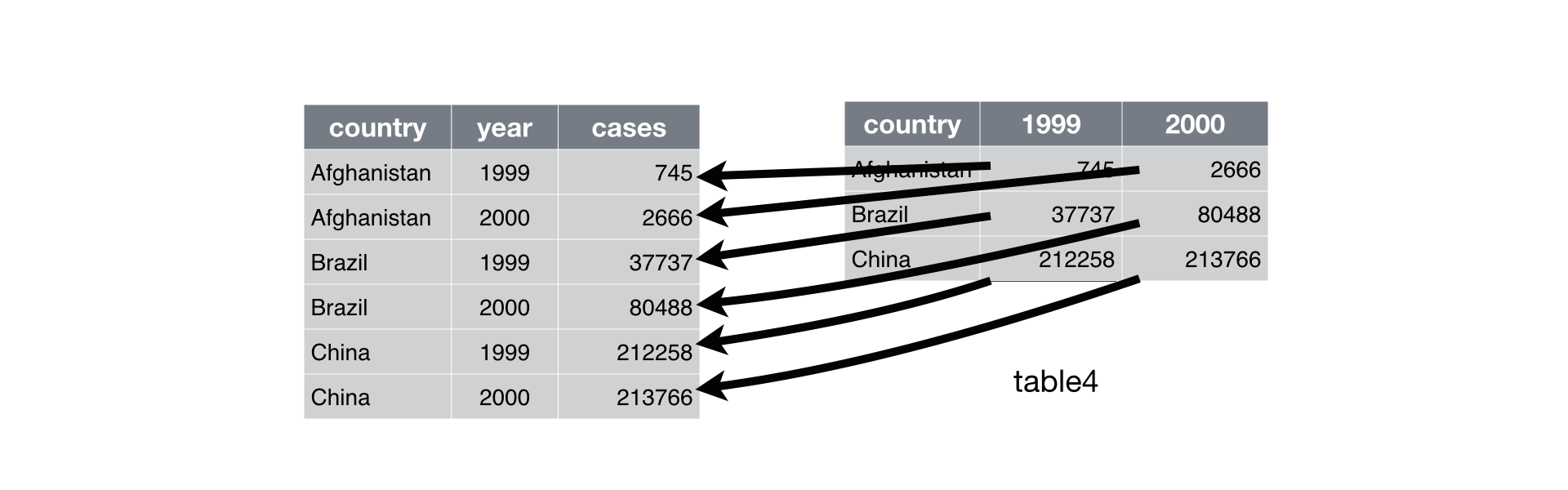
Figure 1: Convert from wide to long. Taken from R for Data Science.
To go from wide to long we use gather to sweep up a set of columns into one
key-value pair. The arguments for gather are:
- The name of a new column that contains the original column names
- The name of a new column that contains the values from the original columns
- The original columns we either want or do not want “gathered” up.
Compare how table4b looks normally and then after converting to the long form
with gather().
# Original data
table4b
#> # A tibble: 3 x 3
#> country `1999` `2000`
#> * <chr> <int> <int>
#> 1 Afghanistan 19987071 20595360
#> 2 Brazil 172006362 174504898
#> 3 China 1272915272 1280428583
# Convert to long form by stacking population by each year
# Use minue to exclude a variable (country) from being "gathered"
table4b %>%
gather(year, population, -country)
#> # A tibble: 6 x 3
#> country year population
#> <chr> <chr> <int>
#> 1 Afghanistan 1999 19987071
#> 2 Brazil 1999 172006362
#> 3 China 1999 1272915272
#> 4 Afghanistan 2000 20595360
#> 5 Brazil 2000 174504898
#> 6 China 2000 1280428583
# This does the same:
table4b %>%
gather(year, population, `1999`, `2000`)
#> # A tibble: 6 x 3
#> country year population
#> <chr> <chr> <int>
#> 1 Afghanistan 1999 19987071
#> 2 Brazil 1999 172006362
#> 3 China 1999 1272915272
#> 4 Afghanistan 2000 20595360
#> 5 Brazil 2000 174504898
#> 6 China 2000 1280428583
This can be challenging to conceptually grasp at first. Let’s try an example
situation when you may use gather(). Let’s say you wanted to find out the
means of several characteristics of a population. You could use the group_by()
and summarise() combination on their own, but another method is using gather()
to group by and summarise all the variables at once.
# Keep only variables of interest
nhanes_chars <- NHANES %>%
select(SurveyYr, Gender, Age, Weight, Height, BMI, BPSysAve)
nhanes_chars
#> # A tibble: 10,000 x 7
#> SurveyYr Gender Age Weight Height BMI BPSysAve
#> <fct> <fct> <int> <dbl> <dbl> <dbl> <int>
#> 1 2009_10 male 34 87.4 165. 32.2 113
#> 2 2009_10 male 34 87.4 165. 32.2 113
#> 3 2009_10 male 34 87.4 165. 32.2 113
#> 4 2009_10 male 4 17 105. 15.3 NA
#> 5 2009_10 female 49 86.7 168. 30.6 112
#> 6 2009_10 male 9 29.8 133. 16.8 86
#> 7 2009_10 male 8 35.2 131. 20.6 107
#> 8 2009_10 female 45 75.7 167. 27.2 118
#> 9 2009_10 female 45 75.7 167. 27.2 118
#> 10 2009_10 female 45 75.7 167. 27.2 118
#> # … with 9,990 more rows
# Convert to long form, excluding year and gender
nhanes_long <- nhanes_chars %>%
gather(Measure, Value, -SurveyYr, -Gender)
nhanes_long
#> # A tibble: 50,000 x 4
#> SurveyYr Gender Measure Value
#> <fct> <fct> <chr> <dbl>
#> 1 2009_10 male Age 34
#> 2 2009_10 male Age 34
#> 3 2009_10 male Age 34
#> 4 2009_10 male Age 4
#> 5 2009_10 female Age 49
#> 6 2009_10 male Age 9
#> 7 2009_10 male Age 8
#> 8 2009_10 female Age 45
#> 9 2009_10 female Age 45
#> 10 2009_10 female Age 45
#> # … with 49,990 more rows
# Calculate mean on each measure, by gender and year
nhanes_long %>%
group_by(SurveyYr, Gender, Measure) %>%
summarise(MeanValue = mean(Value, na.rm = TRUE))
#> # A tibble: 20 x 4
#> # Groups: SurveyYr, Gender [4]
#> SurveyYr Gender Measure MeanValue
#> <fct> <fct> <chr> <dbl>
#> 1 2009_10 female Age 38.0
#> 2 2009_10 female BMI 27.0
#> 3 2009_10 female BPSysAve 116.
#> 4 2009_10 female Height 157.
#> 5 2009_10 female Weight 67.1
#> 6 2009_10 male Age 35.5
#> 7 2009_10 male BMI 26.7
#> 8 2009_10 male BPSysAve 120.
#> 9 2009_10 male Height 168.
#> 10 2009_10 male Weight 76.3
#> 11 2011_12 female Age 37.3
#> 12 2011_12 female BMI 26.5
#> 13 2011_12 female BPSysAve 117.
#> 14 2011_12 female Height 156.
#> 15 2011_12 female Weight 65.3
#> 16 2011_12 male Age 36.2
#> 17 2011_12 male BMI 26.4
#> 18 2011_12 male BPSysAve 120.
#> 19 2011_12 male Height 167.
#> 20 2011_12 male Weight 75.3
Note that summarising by mean can only be done with continuous variables. Anyway, with the long form, you can do more with less code ☺️
spread(): (If time) Converting from long to wide form
You can also convert to wide from long. This is not as commonly done, since most
analyses are better in the long form. But sometimes you may need to have a wide
form. Here you can use spread(), which takes three arguments: 1) the data, 2)
the key column (or column with identifying information), 3) the value
column (the one with the numbers/values). We’ll use a pipe so we can ignore the
data argument.
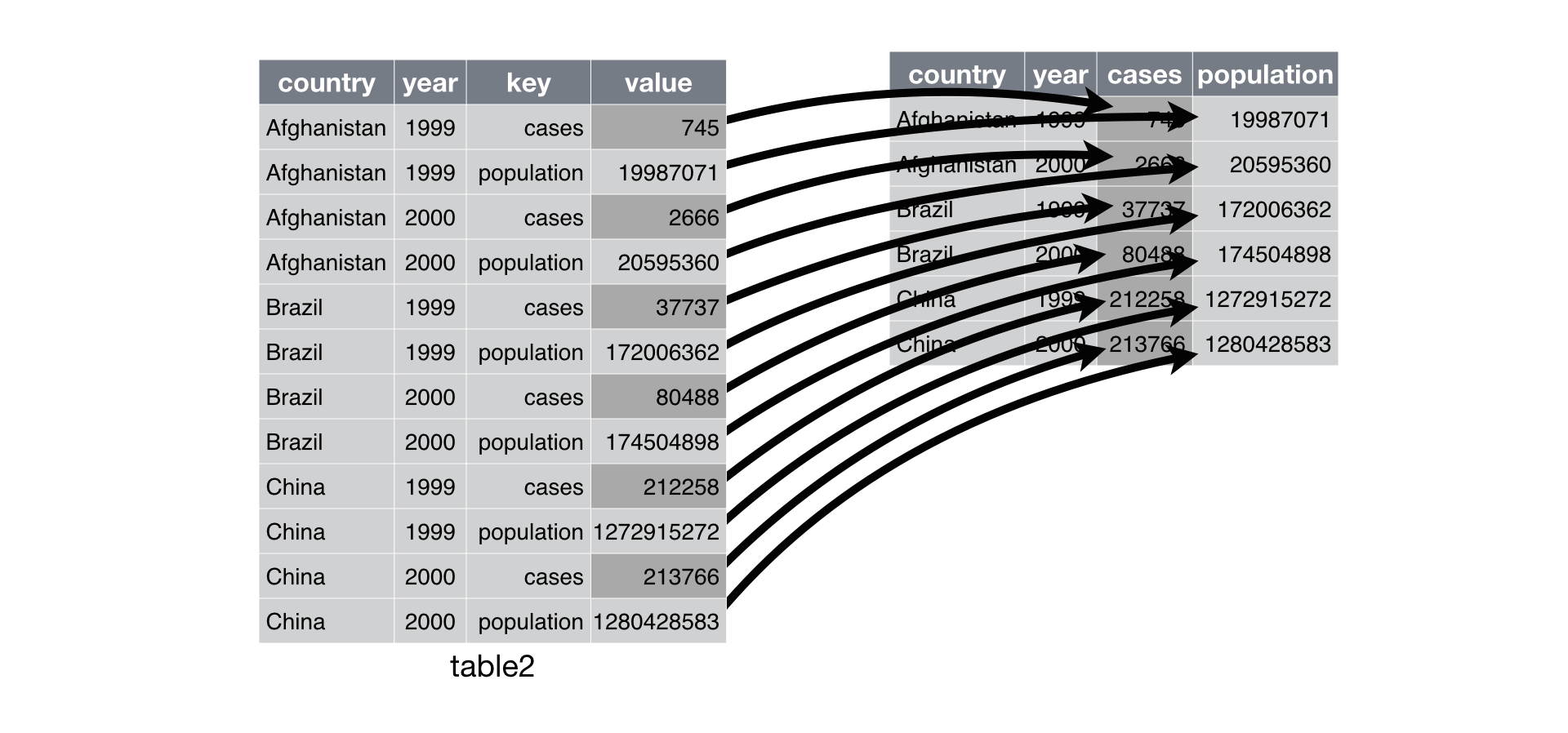
Figure 2: ‘Spreading’ from long to wide. Taken from the R for Data Science book.
# Using a small dataset:
table2
#> # A tibble: 12 x 4
#> country year type count
#> <chr> <int> <chr> <int>
#> 1 Afghanistan 1999 cases 745
#> 2 Afghanistan 1999 population 19987071
#> 3 Afghanistan 2000 cases 2666
#> 4 Afghanistan 2000 population 20595360
#> 5 Brazil 1999 cases 37737
#> 6 Brazil 1999 population 172006362
#> 7 Brazil 2000 cases 80488
#> 8 Brazil 2000 population 174504898
#> 9 China 1999 cases 212258
#> 10 China 1999 population 1272915272
#> 11 China 2000 cases 213766
#> 12 China 2000 population 1280428583
# Convert to wide form
table2 %>%
spread(key = type, value = count)
#> # A tibble: 6 x 4
#> country year cases population
#> <chr> <int> <int> <int>
#> 1 Afghanistan 1999 745 19987071
#> 2 Afghanistan 2000 2666 20595360
#> 3 Brazil 1999 37737 172006362
#> 4 Brazil 2000 80488 174504898
#> 5 China 1999 212258 1272915272
#> 6 China 2000 213766 1280428583
The key is the discrete value that will make up the new column names, while the value will be column that will make up the values of the new columns.
Final exercise: Replicate the output from the given dataset.
Time: Until end of the session.
In the exercise-wrangling.R file, copy the code below to get the data into the
form needed. Using everything you learned in this session, try to recreate the
table below from the dataset given here.
NHANES
#> # A tibble: 10,000 x 76
#> ID SurveyYr Gender Age AgeDecade AgeMonths Race1 Race3 Education
#> <int> <fct> <fct> <int> <fct> <int> <fct> <fct> <fct>
#> 1 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 2 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 3 51624 2009_10 male 34 " 30-39" 409 White <NA> High Sch…
#> 4 51625 2009_10 male 4 " 0-9" 49 Other <NA> <NA>
#> 5 51630 2009_10 female 49 " 40-49" 596 White <NA> Some Col…
#> 6 51638 2009_10 male 9 " 0-9" 115 White <NA> <NA>
#> 7 51646 2009_10 male 8 " 0-9" 101 White <NA> <NA>
#> 8 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 9 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> 10 51647 2009_10 female 45 " 40-49" 541 White <NA> College …
#> # … with 9,990 more rows, and 67 more variables: MaritalStatus <fct>,
#> # HHIncome <fct>, HHIncomeMid <int>, Poverty <dbl>, HomeRooms <int>,
#> # HomeOwn <fct>, Work <fct>, Weight <dbl>, Length <dbl>, HeadCirc <dbl>,
#> # Height <dbl>, BMI <dbl>, BMICatUnder20yrs <fct>, BMI_WHO <fct>,
#> # Pulse <int>, BPSysAve <int>, BPDiaAve <int>, BPSys1 <int>,
#> # BPDia1 <int>, BPSys2 <int>, BPDia2 <int>, BPSys3 <int>, BPDia3 <int>,
#> # Testosterone <dbl>, DirectChol <dbl>, TotChol <dbl>, UrineVol1 <int>,
#> # UrineFlow1 <dbl>, UrineVol2 <int>, UrineFlow2 <dbl>, Diabetes <fct>,
#> # DiabetesAge <int>, HealthGen <fct>, DaysPhysHlthBad <int>,
#> # DaysMentHlthBad <int>, LittleInterest <fct>, Depressed <fct>,
#> # nPregnancies <int>, nBabies <int>, Age1stBaby <int>,
#> # SleepHrsNight <int>, SleepTrouble <fct>, PhysActive <fct>,
#> # PhysActiveDays <int>, TVHrsDay <fct>, CompHrsDay <fct>,
#> # TVHrsDayChild <int>, CompHrsDayChild <int>, Alcohol12PlusYr <fct>,
#> # AlcoholDay <int>, AlcoholYear <int>, SmokeNow <fct>, Smoke100 <fct>,
#> # Smoke100n <fct>, SmokeAge <int>, Marijuana <fct>, AgeFirstMarij <int>,
#> # RegularMarij <fct>, AgeRegMarij <int>, HardDrugs <fct>, SexEver <fct>,
#> # SexAge <int>, SexNumPartnLife <int>, SexNumPartYear <int>,
#> # SameSex <fct>, SexOrientation <fct>, PregnantNow <fct>
| Gender | Measure | 2009_10 | 2011_12 |
|---|---|---|---|
| female | Age | 44.02 | 44.15 |
| female | AgeDiabetesDiagnosis | 48.13 | 46.51 |
| female | BMI | 28.96 | 28.60 |
| female | BPDiaAve | 67.69 | 70.04 |
| female | BPSysAve | 116.04 | 117.90 |
| female | DrinksOfAlcoholInDay | 2.34 | 2.20 |
| female | MoreThan5DaysActive | 0.31 | 0.39 |
| female | NumberOfBabies | 2.44 | 2.31 |
| female | Poverty | 2.93 | 2.88 |
| female | TotalCholesterol | 5.14 | 5.11 |
| male | Age | 43.11 | 43.88 |
| male | AgeDiabetesDiagnosis | 47.58 | 45.14 |
| male | BMI | 28.76 | 28.70 |
| male | BPDiaAve | 70.94 | 73.14 |
| male | BPSysAve | 121.61 | 122.43 |
| male | DrinksOfAlcoholInDay | 3.52 | 3.71 |
| male | MoreThan5DaysActive | 0.29 | 0.36 |
| male | NumberOfBabies | NaN | NaN |
| male | Poverty | 3.01 | 2.97 |
| male | TotalCholesterol | 5.07 | 4.90 |
Resources for learning and help
For learning:
For help:
- Data wrangling cheatsheet
- StackOverflow for tidyr
- StackOverflow for dplyr
- Quick package information using RStudio:
?tidyr,?dplyr
Acknowledgements
Parts of this lesson were modified from a session taught at the Aarhus University Open Coders, with contributions from Elena Dudukina, PhD student at AU in Clinical Epidemiology, as well as from the dplyr sessions of the UofTCoders course. Much inspiration was also taken from the R for Data Science book.